“CRISPR-Cas systems” in bacteria and viruses identify and destroy invading viral sequences. It is bacterial and archaeal immune system for protection against viral infections. In 2012, CRISPR-Cas system was recognised as a genome editing tool. Since then, wide range of CRISPR-Cas systems have been developed and have found applications in areas such as in gene therapy, diagnostics, research and crop improvement. However, currently available CRISPR-Cas systems have limited clinical use due to frequent occurrences of off-target editing, unexpected DNA mutations and inheritable problems. Researchers have recently reported a novel CRISPR-Cas system that can target and destroy mRNA and proteins associated with different genetic diseases more accurately without off-target impact and inheritable problems. Named Craspase, it is the first CRISPR-Cas system that shows protein editing function. It is also the first system that can edit both RNA and protein. Because Craspase overcomes many limitations of existing CRISPR-Cas systems, it has potential to revolutionise gene therapy, diagnostics and monitoring, biomedical research, and crop improvement.
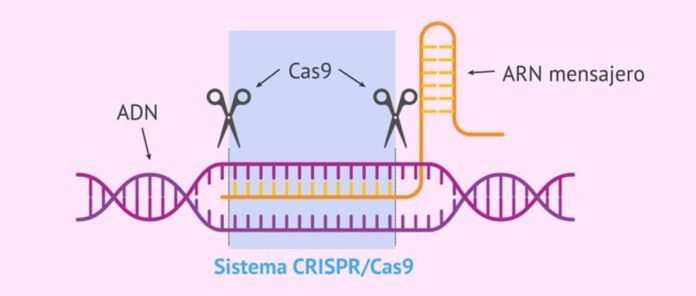
“CRISPR-Cas system” is natural immune system of bacteria and archaea against viral infections that identifies, binds and degrades the sequences in the viral gene to protect. It consists of two parts – bacterial RNA transcribed from the viral gene incorporated in the bacterial genome after first infection (called CRISPR, this identifies the target sequences of the invading viral genes) and an associated destroyer protein called “CRISPR associated protein (Cas)” which binds and degrades the identified sequences in the viral gene to protect the bacteria against viruses.
CRISPER stands for “clustered regularly interspaced short palindromic repeats”. It is transcribed bacterial RNA characterised by palindromic repeats.
Palindromic repeats (CRISPRs) were first discovered in the sequences of E. coli in 1987. In 1995, Francisco Mojica observed similar structures in archaea, and it was he who first thought of these as a part of the immune system of bacteria and archaea. In 2008, it was experimentally demonstrated for the first time that the target of the immune system of bacteria and archaea was foreign DNA and not mRNA. The mechanism of identification and degradation viral sequences suggested that such systems could be used as a tool for genome editing. Since its recognition as a genome editing tool in 2012, CRISPR–Cas system has come a very long way as a firmly established standard gene editing system and has found a wide range of applications in biomedicine, agriculture, pharmaceutical industries including in clinical gene therapy1,2.
A wide range of CRISPR-Cas systems are already identified and currently available for monitoring and editing DNA/RNA sequences for research, drug screening, diagnostics and treatments. The current CRISPR/Cas systems are divided into 2 classes (Class 1 and 2) and six types (Type I to XI). Class 1 systems have multiple Cas proteins which need to form a functional complex to bind and act on their targets. On the other hand, Class 2 systems have only one large Cas protein for binding and degrading target sequences which makes Class 2 systems easier to use. Commonly used Class 2 systems are Cas 9 Type II, Cas13 Type VI, and Cas12 Type V. These systems may have undesired collateral effects I.e., off-target impact and cytotoxicity3,5.
Gene therapies based on current CRISPR- Cas systems have limited clinical use because of frequent occurrences of off-target editing, unexpected DNA mutations, including big DNA fragment deletions and large DNA structural variants at both on-target and off-target sites that leads to cell deaths and other inheritable problems.
Craspase (or CRISPR-guided caspase)
Researchers have recently reported a novel CRISPER-Cas system which is a Class 2 Type III-E Cas7-11 system associated with a caspase-like protein hence named Craspase or CRISPR-guided caspase 5 (Caspases are cysteine proteases that play key role in apoptosis in breaking down cellular structures). It has potential applications in areas like gene therapy and diagnostics. Craspase is RNA-guided and RNA-targeted and do not get involved with the DNA sequences. It can target and destroy mRNA and proteins associated with different genetic diseases more accurately without off-target impact. Thus, elimination of genes associated with diseases is possible by cleavage at mRNA or protein level. Also, when linked with specific enzyme, Craspase can also be used to modify functions of proteins. When its RNase and protease functions are removed, Craspase becomes deactivated (dCraspase). It has no cutting function but binds with RNA and protein sequences. Therefore, dCraspase can be used in diagnostics and imaging to monitor and diagnose diseases or viruses.
Craspase is the first CRISPR-Cas system that shows protein editing function. It is also the first system that can edit both RNA and protein. Its gene editing function comes at minimal off-target effects and no inheritable problems. Hence, Craspase is likely to be safer in clinical use and therapeutics than other currently available CRISPR- Cas systems 4,5.
Because Craspase overcomes many limitations of existing CRISPR-Cas systems, it has potential to revolutionise gene therapy, diagnostics and monitoring, biomedical research, and crop improvement. More research is needed to develop reliable delivery system to precisely target disease causing genes in the cells before proving safety and efficacy in clinical trials.
***
References:
- Gostimskaya, I. CRISPR–Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochemistry Moscow 87, 777–788 (2022). https://doi.org/10.1134/S0006297922080090
- Chao Li et al 2022. Computational Tools and Resources for CRISPR/Cas Genome Editing. Genomics, Proteomics & Bioinformatics. Available online 24 March 2022. DOI: https://doi.org/10.1016/j.gpb.2022.02.006
- van Beljouw, S.P.B., Sanders, J., Rodríguez-Molina, A. et al. RNA-targeting CRISPR–Cas systems. Nat Rev Microbiol 21, 21–34 (2023). https://doi.org/10.1038/s41579-022-00793-y
- Chunyi Hu et al 2022. Craspase is a CRISPR RNA-guided, RNA-activated protease. Science. 25 Aug 2022. Vol 377, Issue 6612. pp. 1278-1285. DOI: https://doi.org/10.1126/science.add5064
- Huo, G., Shepherd, J. & Pan, X. Craspase: A novel CRISPR/Cas dual gene editor. Functional & Integrative Genomics 23, 98 (2023). Published: 23 March 2023. DOI: https://doi.org/10.1007/s10142-023-01024-0
***